Looking back, it's easy to forget the significance of IBM Watson's Jeopardy win in February 2011. At that time, iPhones had no built-in Siri, deep learning was still obscure, and few people would believe how pervasive devices such as Google Home and Amazon Alexa have now become. Watson's win made artificial intelligence advances tangible for millions of viewers.
Yet despite this breakthrough, for many biomedical researchers, it's still 2011. While IBM quickly applied Watson in healthcare, its efforts (especially relative to the hype) have been largely unsuccessful. And while several companies now do successfully apply machine learning to identifying drug candidates, they don't address the many inefficiencies that can make developing those candidates so challenging, expensive and frustrating.
But this is starting to change. At BenchSci, where we apply machine learning to antibody selection and reagent intelligence for drug discovery, we've seen this first hand. And in my research and conversations with related startups, it seems we're not alone. Rather, if phase 1 of artificial intelligence in drug discovery was Watson and phase 2 was the launch of drug candidate-identification startups, phase 3 seems to be growth in the "long tail"—companies using machine learning in highly specific ways to streamline and improve the many day-to-day biomedical research tasks required to bring those candidates to market.
The 3Ds Powering AI in Drug Discovery: Domain Expertise, Deep Learning and Data
What's driving this trend is domain expertise, deep learning and data.
Let's start with domain expertise. In my experience with IBM Watson within a pharma company, IBM struggles to articulate use cases. Since they lack the domain expertise, they look to partners to identify and volunteer biomedical problems to solve, while they bring the underlying technology. This approach might make sense if Watson were truly unique, but from a technical perspective, under the hood, "Watson" is an assemblage of software tools—many acquired by IBM and mashed together, most available elsewhere (including as open source software) and some simply rebranded (such as IBM Watson Campaign Automation for marketing, formerly Silverpop).
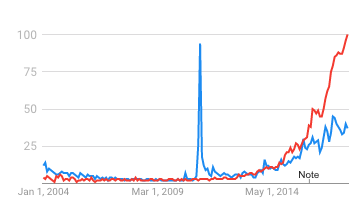 This brings us to the second trend: deep learning. While Watson's Jeopardy victory had high visibility—and truly was a breakthrough—a less public triumph the following year was the successful application of deep learning to the ImageNet image recognition contest, dramatically dropping the error rate from 25.8% to 16.4% (and by 2015 to greater than human performance). This sparked a gold rush to apply deep learning (which was invented in the 1980s, but which lacked sufficient computational power and data for practical applications) to multiple industries, including healthcare. Notably, Watson did not apply deep learning in its Jeopardy-winning architecture, and it wasn't until 2015 that IBM would add the capability to the commercial version, falling behind leaders such as Google with DeepMind. As shown in the Google Trends chart above, interest in "deep learning" (red) rapidly overtook interest in "IBM Watson" (blue).
This brings us to the second trend: deep learning. While Watson's Jeopardy victory had high visibility—and truly was a breakthrough—a less public triumph the following year was the successful application of deep learning to the ImageNet image recognition contest, dramatically dropping the error rate from 25.8% to 16.4% (and by 2015 to greater than human performance). This sparked a gold rush to apply deep learning (which was invented in the 1980s, but which lacked sufficient computational power and data for practical applications) to multiple industries, including healthcare. Notably, Watson did not apply deep learning in its Jeopardy-winning architecture, and it wasn't until 2015 that IBM would add the capability to the commercial version, falling behind leaders such as Google with DeepMind. As shown in the Google Trends chart above, interest in "deep learning" (red) rapidly overtook interest in "IBM Watson" (blue).
Meanwhile, companies that were built around deep learning began procuring and analyzing massive amounts of data—the third ingredient. This has been fuelled by growth in sources of data, including increased access to published research, electronic health records, the various "omics" fields and even social media.
The result is that there are now at least 11 companies that combine domain expertise, deep learning and data to identify drug candidates: Atomwise, BenevolentAI, Cloud, Exscienta, Insilico Medicine, NuMedii, Numerate, Qrativ, Recursion, TwoXAR, and Verge.
But this isn't where the trend ends.
From Swiss Army Knife Machine Learning to Scalpels
Just as the mix of domain expertise, deep learning and data gave drug candidate identification companies an advantage over generalists such as IBM, they give advantage to companies that focus on niche aspects of the drug discovery and development process that are currently underserved. The result is growth in companies such as BenchSci that use machine learning to focus on very specific aspects of healthcare and drug discovery—the long tail.
In February, for example, research company CB Insights highlighted 106 healthcare startups using machine learning, up from 20 in 2012 and 70 just last year. Their list is now up to 176. Such lists now include niche applications of artificial intelligence in healthcare such as Envisagenics for RNA splicing analytics, Celldom for automated single cell analysis and Deep 6 AI for finding patients for clinical trials faster.
From what we're seeing in the market—the feedback we get from customers and users—there is unmet demand for such specific use cases throughout pharmaceutical and biotechnology companies. These solutions need to be targeted to specific users, tuned to specific use cases, trained by domain experts, and built with deep data addressing a specific niche—the more general use case of identifying drug candidates is already crowded.
"The AI approach has to be very specific to the problem at hand," agrees Deep 6 CEO Wout Brusselaers. " It also needs to be augmented by human user workflow to validate algorithm-derived results and offer users a delightful experience overall. We are big believers in synergistic relationships between artificial and human intelligence."
So if you're a lab manager or research director looking at the machine learning landscape, it might be worth identifying the specific pain points in your pipeline and looking for best-in-class solutions tailored to address them. And if you're a startup looking to apply machine learning in drug discovery, take heart: while there are big players at the head of the tail, even IBM struggled there, and there's plenty of long tail for the taking.
Image credit: IBM

