At BenchSci we’re committed to making science better, faster, and more impactful. Our mission is to exponentially increase the speed and quality of life-saving research. And we do so by empowering scientists with the world’s most advanced biomedical artificial intelligence so they can run more successful experiments.
Today we’re pleased to announce a significant step forward in achieving that mission by introducing RNA interference (RNAi) products on our new AI-Assisted Reagent Selection platform.
A few months back we shared that we’d expanded our ML-based analyses to decode the usage of additional reagents in the published literature, and added antibody validation.
Now, we’re officially moving beyond antibodies, enabling you to search for additional reagents integral to the preclinical scientific pipeline. Our goal is to reduce the effort you have to spend finding reagents used in the literature and validating them. Today we’re making RNAi data available, and there are more reagents coming soon. Currently this product is only available to enterprise customers, while the existing AI-Assisted Antibody Selection platform remains free for academic researchers, and always will.
BenchSci & RNAi
Our AI is trained to identify knockout and knockdown experiments, intelligently link the data to vendor catalog numbers found throughout the text, or distill the RNAi nucleotide sequences themselves (currently a beta feature, read more about it here) from custom or in-house nucleotide synthesis. Generating bespoke RNAi reagents is relatively easy these days, which means that having access to sequence information is just as, if not more, useful than a vendor catalog number in terms of informing your experiments.
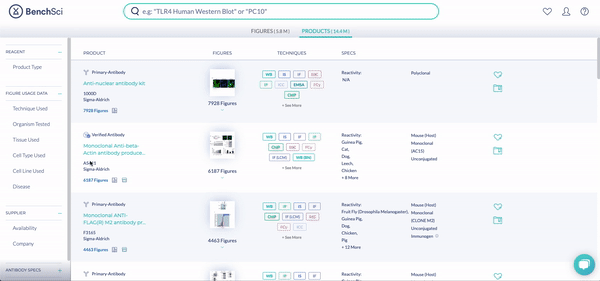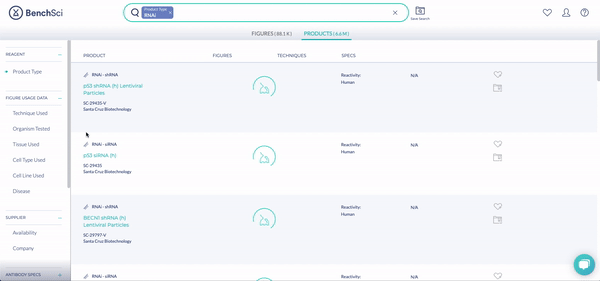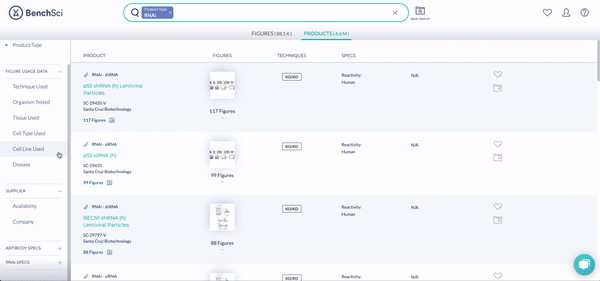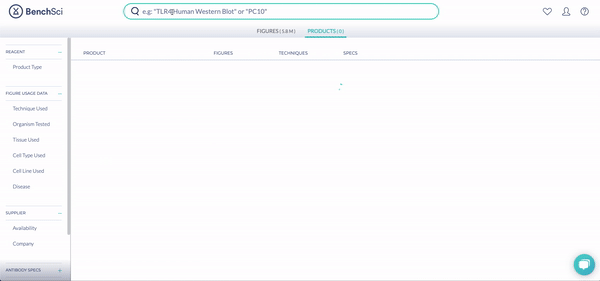
In the research pipeline, antibodies and RNA interference experiments are often closely associated, where one can verify the efficacy of the other. Some of the challenges you face with designing RNAi experiments will be familiar from working with antibodies. Finding a product or sequence that effectively targets the intended gene/target for your specific experiment with supporting data is particularly difficult because modern search platforms are not optimized to recognize RNAi nucleotides. Not to mention that the numerous protein naming conventions and abbreviations can complicate finding a comprehensive list of results for your target. To address these challenges, on BenchSci advanced bioinformatics and ontologies connect RNAi experiments, figures from the literature, product specifications, and nucleotide sequences. This means that you can search our database and hone-in on products and figures from the literature that meet highly specific criteria, like lentiviral-delivered shRNA used to knockdown or knockout BRCA1.
RNAi products feature a new icon (see above!). You can display exclusively RNAi using the new filter, Product Type (see below). The options are Antibody and RNAi products, with subcategories for siRNA (small interfering RNA), shRNA (short hairpin RNA), and miRNA (micro RNA). We are collaborating with 25 vendors to give you access to more than 6.6 million RNAi products at launch, filterable by 10 product and vendor specifications tailored for RNAi.
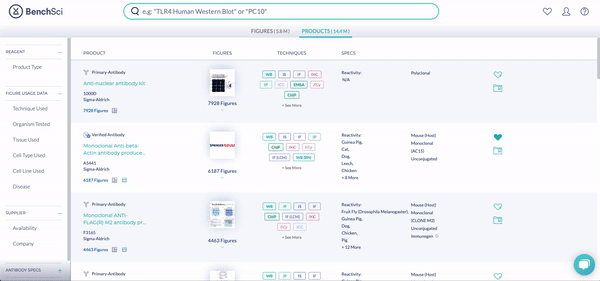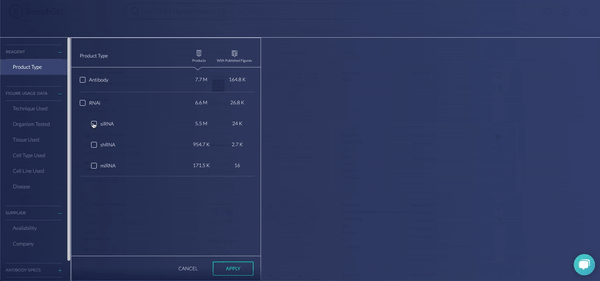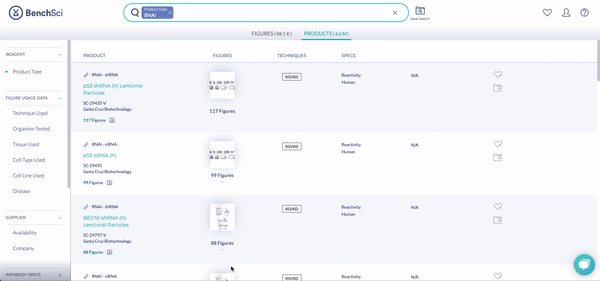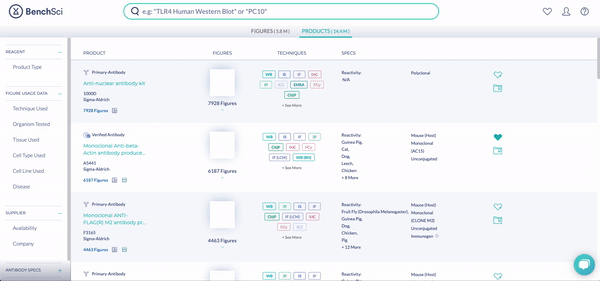
RNAi Specs is a growing category of new filters specifically designed for RNAi that you can use to better define what you’re looking for (see below): whether a double stranded oligo or lentiviral plasmid (Format), Puromycin and/or Kanamycin Selectable Markers, with Reactivity in Humans or Rhesus Monkey.
The RNAi platform is under active development as we work hard to expand our features, like the number of filterable specifications, vendors, products, and figures. We would love to hear about how you search for RNAi products or what (new) filters would get the most use from you - share your thoughts here.
We’re proud to announce this new product, and look forward to continuing to support you and your research efforts in the future. Let us know what you think in the comments!
