At BenchSci, we constantly speak with fellow scientists to understand how we can further improve the platform, both in usability and data coverage.
We really appreciate their feedback and support, and would like to showcase their research and profiles in our "Researcher Highlight" series.
For our first highlight, I chatted with Ladan Gheiratmand, a postdoctoral fellow at the Mount Sinai Hospital in Toronto who loves microscopy and Zumba!
Can you tell us your educational background and why you decided to come to Toronto for your research career?
I did my PhD at the National University of Singapore and now I'm a postdoc in Dr. Laurence Pelletier's lab at the Lunenfeld-Tanenbaum Research Institute in Toronto.
I joined Dr. Pelletier's lab because I always had a passion for centriole and cilia biology and I love how Toronto is very multicultural. I love everything about Toronto except for its winter!
Another reason why I joined this lab is because of its amazing microscopy facility. I really really like microscopy and looking at fluorescent images. I can do microscopy for hours!
What project are you currently working on?
I'm studying the proteome of centriolar satellites (CSs), which are these little electron-dense granules that surround the centrioles. The composition and function of CSs are not very well known. There are studies that have shown that ciliogenesis, or cilia generation, is impaired by the knockdown of CSs. There are also mutations in the genes of CSs that are associated with ciliopathies, or diseases related to cilia.
So the question my study tries to answer is, what is the protein composition and function of CSs? My idea is that CSs are acting like an organelle themselves, meaning together these granules have a physiological function, rather than individual proteins by itself.
My project focuses on the proteomic analysis of CSs; I've done BioID, a proximity-based proteomics method, to screen the satellites that interact with each other, and then following up on the biogenesis using knock-down/out. Using this method, I've identified 9 of the 30 CSs that are currently known, published in Cell in 2015.
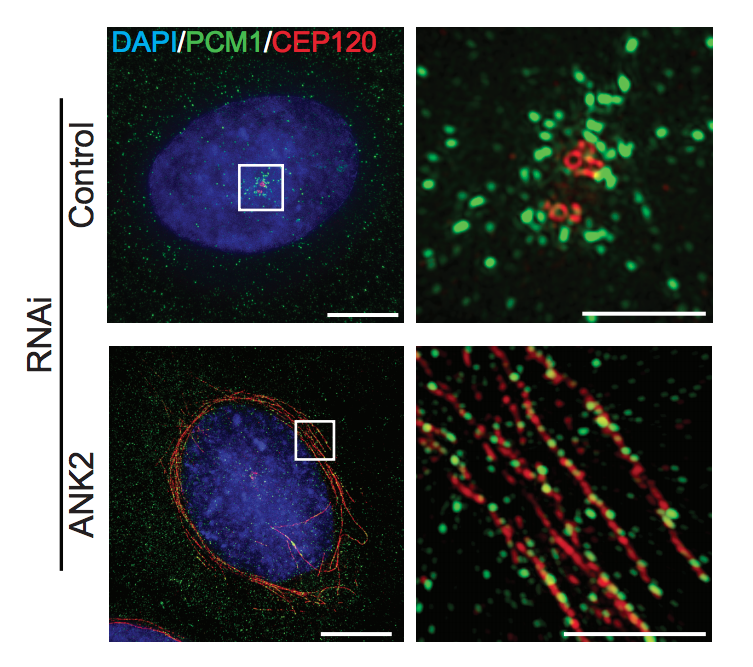
Figure S5B: ANK2 knockdown reduced ciliation and perturbed satellite morphology
That is very impressive! Do you have any tips or advices for younger generation of scientists?
I have mentored many students, and the one thing I always try to teach them is: Understand why you are doing an experiment before you start. Keep asking yourself: What questions am I trying to answer?
A lot of times, as we delve into a project, it becomes a daily routine and we forget the question behind why we are tagging this and transfecting that. Reminding yourself the purpose of your project from time to time will help you to be more creative with your approach, as well as to help you troubleshoot for errors.
Another thing that is really important—which I am actually pretty bad at!—is to be very organized with your data. I enjoy working at the bench much more than in front of a computer. I often find myself having so much data that I haven’t analyzed before I moved on to the next experiment, only to find out later that I didn’t really need to do the additional experiments because I already had the data I needed.
I would encourage young scientists to develop a system for data organization early on and make it a good habit on their research path.
How has BenchSci helped with your research?
Because the nature of my studies is very exploratory, I'm always searching for antibodies that can bind to candidate proteins with high specificity to check the localizations or to confirm my knock-down/out.
Before I knew about BenchSci, I used to spend at least 2-3 hours per protein to go over all the companies, the antibodies, and the papers just to find out what are my options. I used to have so many word files for each protein documenting my research on antibodies. And then I have to go through them to find out which of the available antibodies have been published in the applications and cell lines that I plan to use.
When I heard about BenchSci, I say to myself: This is exactly what I needed!
If you guys hadn't built the platform, I probably would have tried it myself! It saves so much time for me because I'm always looking at several proteins per study (30-40 proteins so far). I can choose from the list of candidate proteins the ones with good antibodies and include those in my experiments.
I also find BenchSci very useful in helping me identify papers that I otherwise wouldn't have known about. Sometimes I go on the platform just to examine published data.
What is your passion outside of the lab?
I enjoy hiking, mountain climbing, cooking—and I love dancing! I've been taking Zumba classes for a long time.
I also love travelling, and I have travelled to about 30 countries so far.
I hope you enjoyed meeting Ladan (virtually!) as much as I did. It's always a great feeling to meet an enthusiastic scientist like Ladan.
If you're also intrigued by her research, here are a couple more papers for you; handpicked by Ladan herself!
- Kofyman et al., (2011), "Structure of Trypanosoma brucei flagellum accounts for its bihelical motion." PNAS
- Gheiratmand et al., (2013), "Biochemical characterization of the bi-lobe reveals a continuous structural network linking the bi-lobe to other single-copied organelles in Trypanosoma brucei." J. Biol. Chem
Are you in love with your research? Let us know in the comment section. We would love to feature your research in our next article!